Chapter 6 Common output for fits
This is a brief summary of common outputs for model fits. The output are shown with the following simple fit:
dat <- cumsum(rnorm(100,0,0.5)) + rnorm(100,0,0.5)
fit <- MARSS(dat)Use the broom package to get the model output in tidy form. The confidence intervals shown are approximate and based on the estimated Hessian matrix. See ?tidy.marssMLE for information on how to change to a different type of confidence interval.
broom::tidy(fit) term estimate std.error conf.low conf.up
1 R.R 0.20232993 0.07256917 0.06009696 0.34456290
2 U.U -0.03710875 0.05988268 -0.15447663 0.08025914
3 Q.Q 0.35208044 0.10659375 0.14316053 0.56100035
4 x0.x0 0.66568193 0.70913982 -0.72420657 2.05557043The get the estimated states use:
head(stats::tsSmooth(fit, type="xtT")) .rownames t .estimate .se
1 X.Y1 1 0.6287215 0.3194423
2 X.Y1 2 0.6361536 0.3325886
3 X.Y1 3 0.1632734 0.3336694
4 X.Y1 4 0.1711874 0.3337600
5 X.Y1 5 0.3302335 0.3337676
6 X.Y1 6 0.2024342 0.3337682Note that generic rownames were given since none were specified for dat.
You can also get all the Kalman filter and smoother estimates for \(\mathbf{x}\) from MARSSkf(). See ?MARSSkf.
There are two types of fitted values that are used in the state-space literature: the one-step-ahead which uses on the data up to \(t-1\) and the smoothed fitted values which uses all the data. Read up on fitted values for MARSS models at ?fitted.marssMLE.
To get the fitted values, the estimated \(\mathbf{y}\), using all the data:
head(fitted(fit, type="ytT")) .rownames t y .fitted
1 Y1 1 0.60321037 0.6287215
2 Y1 2 0.91217455 0.6361536
3 Y1 3 -0.11302441 0.1632734
4 Y1 4 0.08433636 0.1711874
5 Y1 5 0.49507490 0.3302335
6 Y1 6 0.36857168 0.2024342To get the fitted values, the estimated \(\mathbf{y}\), using the data up to time \(t-1\):
head(fitted(fit, type="ytt1")) .rownames t y .fitted
1 Y1 1 0.60321037 0.6285732
2 Y1 2 0.91217455 0.5753577
3 Y1 3 -0.11302441 0.7752737
4 Y1 4 0.08433636 0.1078043
5 Y1 5 0.49507490 0.0540304
6 Y1 6 0.36857168 0.3301366Note, this is the default for fitted(fit).
To make some generic plots and diagnostic plots, you can use autoplot() in the ggplot2 package:
ggplot2::autoplot(fit, plot.type = "xtT")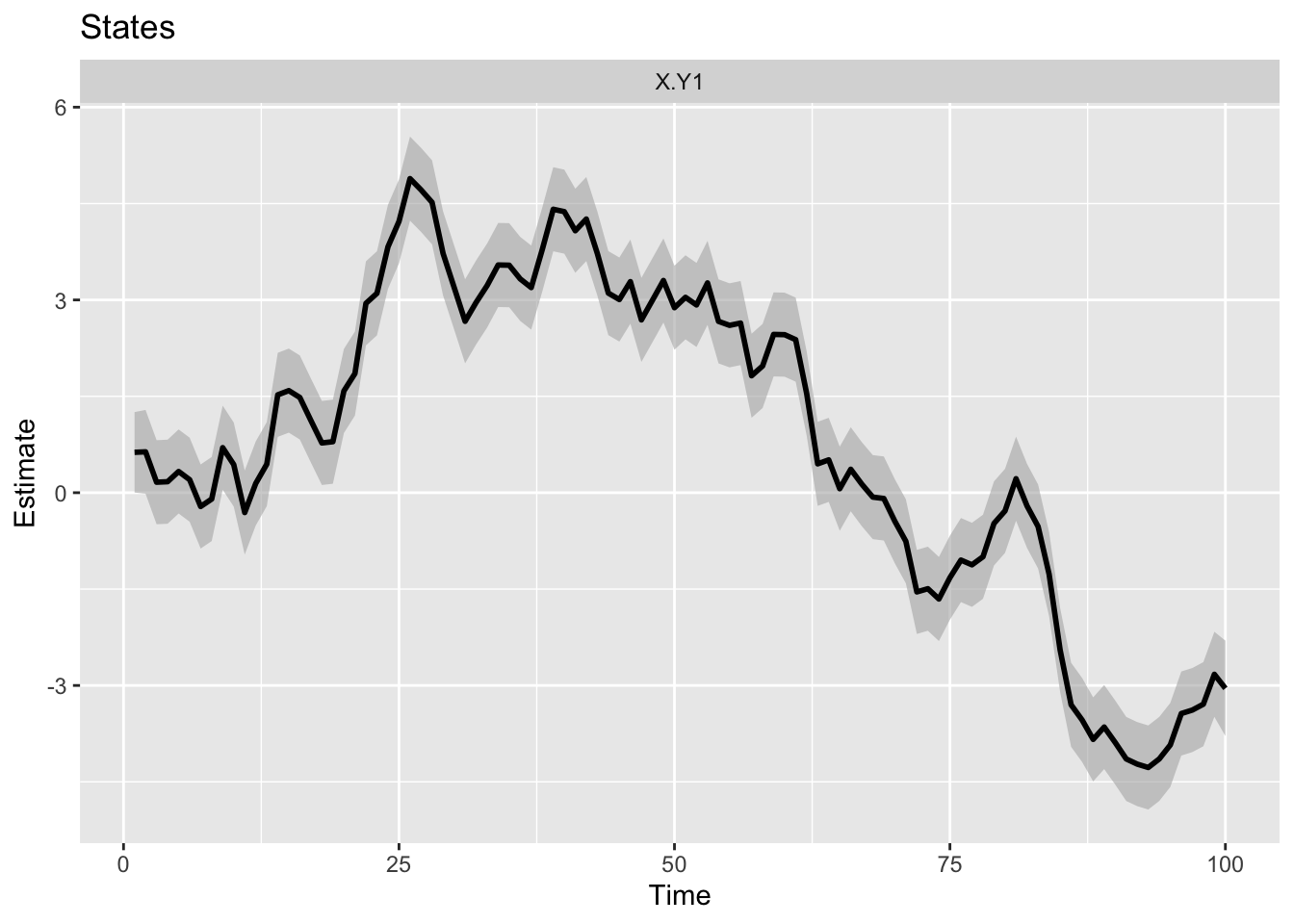
To get residuals, use:
resids <- residuals(fit)
head(format(resids, digits = 2)) type .rownames name t value .fitted .resids .sigma .std.resids
1 ytt1 Y1 model 1 0.6032 0.6286 -0.0254 0.74 -0.0341
2 ytt1 Y1 model 2 0.9122 0.5754 0.3368 0.83 0.4076
3 ytt1 Y1 model 3 -0.1130 0.7753 -0.8883 0.83 -1.0642
4 ytt1 Y1 model 4 0.0843 0.1078 -0.0235 0.84 -0.0281
5 ytt1 Y1 model 5 0.4951 0.0540 0.4410 0.84 0.5279
6 ytt1 Y1 model 6 0.3686 0.3301 0.0384 0.84 0.0460There are many types of residuals possible for MARSS models. The default residuals(fit) will return the innovations residuals, which are what are typically used for residuals diagnostics for state-space models. However residuals() and the companion MARSSresiduals() will return all possible residuals for your model. Read about MARSS residuals in ?residuals.marssMLE.
To get your parameter estimates, use:
coef(fit)Type ?coef.marssMLE to see the different forms that you can get your estimated parameters in. A common form is as a matrix. To get that use coef(fit, type="matrix").
coef(fit, type = "matrix")$Q [,1]
[1,] 0.3520804